AntioxidantDB Summary
Antioxidants are important for maintaining the appropriate balance between the production and the removal of free radicals in the body and thus preventing oxidative stress. The aim of this study was to establish an in vitro antioxidant database. Based on this database, quantitative structure-activity relationship (QSAR) models between the ability of scavenging DPPH free radicals and structural features were established to predict the antioxidant activity of the molecules. The data were curated from the PubChem database and the research articles. To further reduce noise, inorganics, repeats, peptides and molecules with molar mass more than 1000 g/mol were removed. Molecular descriptors were calculated by Mordred and MOE from optimized molecular structure and redundant descriptors were removed by removing collinearity analysis and PCA analysis. Self-organizing map (SOM) algorithms were used for creating the training and test sets in both set of descriptors. The QSAR models were established using random forest (RF) and support vector regression (SVR) two machine learning methods. Distinguishable from other studies, in our work we have interpreted the developed models based on partial dependence plots.
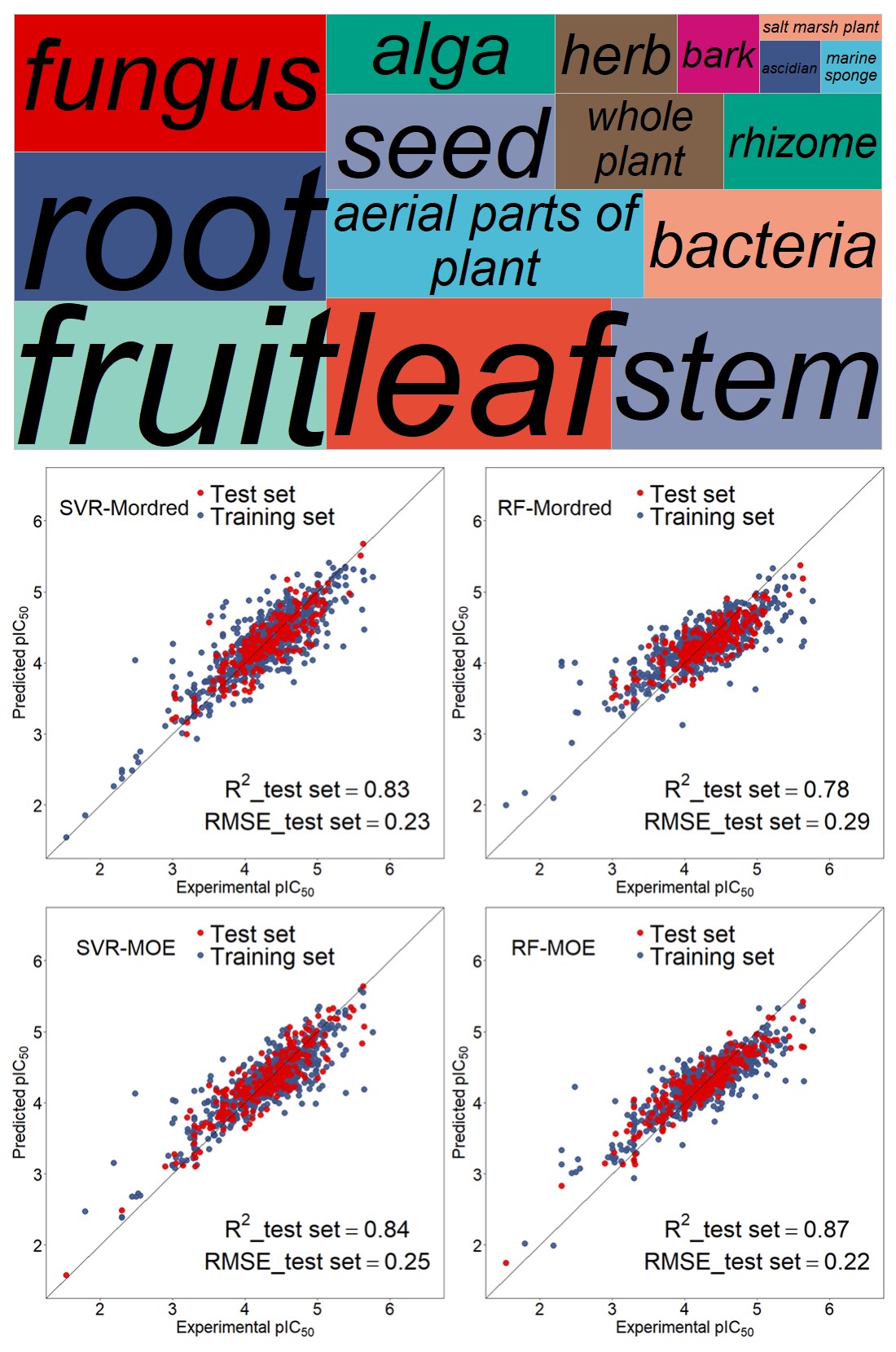
The established antioxidant database AntioxiantDB includes 1008 antioxidant active molecules integrating multidimensional aspects of antioxidant molecules, antioxidant capacity and details of natural source. Data-driven studies based on AntioxidantDB can pave the way for an improved understanding of antioxidant mechanisms. The RF-Mordred model has a relatively weaker result for the internal10-fold cross-validation compared to other models (P<0.05). The RF-MOE was found to be strongly predictive with 0.77±0.06 and 0.27±0.03 for Q2 and RMSECV for the internal validation. Furthermore, the RF-MOE model yielded considerably lower errors (R2=0.87, RMSE=0.22) for external validation. Chemical domains of applicability were defined for both models confirming their reliability and robustness. The study identifies the essential physicochemical descriptors including PEOE_VSA-3, a_don, h_pKa, etc. that effectively contribute in the antioxidant activity. The developed models show sufficient predictive abilities for the screening of virtual libraries for new potential antioxidants.